Help Home > Alignment Search > Running an Alignment Search > Specifying Search Criteria
The Alignment Query Page: Specifying Search Criteria
Alignment Programs
Alignment searches may be run using a variety of programs which compare DNA, protein, or translated nucleotide sequences. These programs are based on three families of alignment algorithms, NCBI's BLAST, Webb Miller's Blastz , and Jim Kent's BLAT (BLAST-Like Alignment Tool).
For more information about the difference between the two algorithms and their variations see:
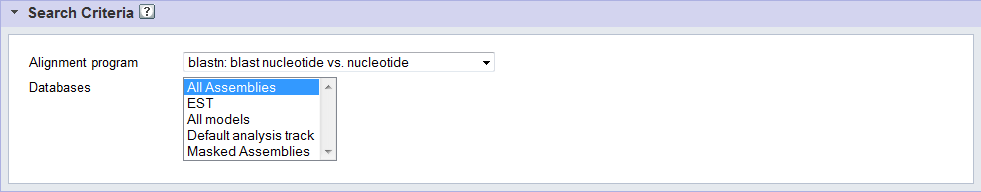
Databases

With each of our organism sites we provide a number of target databases for alignment search which may include:
- All Assemblies available for the organism
- ESTs
- All Models: protein and transcript files for predicted gene models in all model tracks
- Default analysis track: protein and transcript files for predicted gene models in the default analysis track(e.g. GeneCatalog or FilteredModels)
- Masked Assemblies: Repeat-masked assemblies for the organism