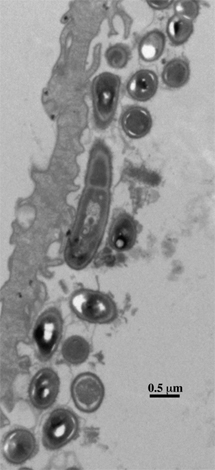
Background L. reuteri 100-23 is a well-characterized rodent-specific strain that has been used in studies concerning the molecular basis of autochthony. Strain 100-23 forms a biofilm on the epithelial surface of the murine forestomach, and has been shown to participate in horizontal gene transfer in the gut. Furthermore, strain 100-23 is readily transformable at high levels by electroporation and can express heterologous genes, which are properties essential for post-genome sequence experimentation. Microbial ecological studies of strain 100-23 have been carried out using a unique mouse colony that we have maintained since the 1980s. The mice in this colony harbour a complex gut microbiota equivalent to their conventional counterparts except that lactobacilli, normally present in the murine gut, are absent. This Lactobacillus -free mouse colony is maintained in isolators by gnotobiotic technology and the animals provide a defined microbiological system with which to compare the colonization ability of lactobacilli as well as Lactobacillus -host interactions. Taxonomic details Relevance to DOE
mission Publications
relating to L. reuteri strain 100-23 Heng, N. C. K., H. F. Jenkinson, and G. W. Tannock. 1997. Cloning and expression of an endo-1,3-1,4- b -glucanase gene from Bacillus macerans in Lactobacillus reuteri . Appl. Environ. Microbiol. 63:3336-3340. McConnell, M.A., A. A. Mercer, and G. W. Tannock. 1991. Transfer of plasmid pAMß1 between members of the normal microflora inhabiting the murine digestive tract and modification of the plasmid in a Lactobacillus reuteri host. Microb. Ecol. Hlth. Dis. 4:343-35. Tannock, G. W., C. Crichton, G. W. Welling, J. P. Koopman, and T. Midtvedt. 1988. Reconstitution of the gastrointestinal microflora of lactobacillus-free mice . Appl. Environ. Microbiol. 54:2971-2975. Tannock, G. W., Ghazally, S., Walter, J., Loach, D., Brooks, H., Cook, G., Surette, M., Simmers, C., Bremer, P., Dal Bello, F., and Hertel, C. 2005. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl. Environ. Microbiol. 71:8419-8425. Walter, J., Chagnaud, P., Tannock, G. W., Loach, D. M., Dal Bello, F.,
Jenkinson, H. F., Hammes, W. P., and Hertel, C. 2005. A high-molecular-mass
surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute
to the ecological performance of Lactobacillus reuteri in the
gut of mice. Appl. Environ. Microbiol. 71:979-986 . Wesney, E., and G. W. Tannock. 1979. Association of rat, pig and fowl biotypes of lactobacilli with the stomach of gnotobiotic mice. Microb. Ecol. 5:35-42. Figure legend.
|
||
|
||
Lactobacillus reuteri 100-23