| |
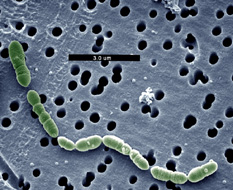
Photo: Jeff Broadbent, Utah State University |
Oenococcus oeni (formerly
called Leuconostoc oenos) is a lactic acid bacterium (LAB) that
occurs naturally in fruit mashes and related habitats. O. oeni is
employed in wineries to carry out the malolactic conversion, an important
secondary fermentation in the production of wine. O. oeni, a facultative
anaerobe, is one of the most acid- and alcohol tolerant LAB.
On the basis of
16S rRNA sequence, Dicks et al. (2) proposed a reclassification of
Leuconostoc oenos into a new genus, Oenococcus oeni. O. oeni shares
relatively little DNA homology with the other genera in the Leuconostoc
branch of the LAB (1). Woese argued that O. oeni is an example
of a fast-evolving microorganism (7). Subsequent comparisons of RNA
polymerase genes contradicted this view (5). Whole genomic comparisons
between O. oeni and the nonacidophilic Leuconostoc mesenteroides (also
sequenced as part of the LABGC project) will help resolve this issue
and shed light on the evolution of this unusual branch.
Perhaps the most
studied aspect of O. oeni is the ability to convert malate to
lactate (the malolactic conversion). This involves uptake of malate,
its decarboxylation to L-lactic acid and CO2, and subsequent export
of end products. The malolactic conversion generates energy for the
cell in the form of a proton motive force (6). Recently the genes encoding
the malate decarboxylase and malate permease have been cloned and characterized
(3, 4).
Most likely due
to difficulties in gene transfer, there are few studies employing genetic
strategies to examine O. oeni. Several bacteriophages and plasmids
have been characterized (6). In addition, the broad host range transposon
Tn916 has been mobilized into the O. oeni genome (8). To date,
however, few chromosomal genes from O. oeni have been sequenced
(< 20 entries in GenBank).
Many researchers
have examined the diversity of O. oeni strains within and around
wineries. Studies have employed various molecular typing methods (protein
profiling, plasmid profiling, RAPD, PFGE, rDNA RFLP, etc.) to discern
regional differences in strains. An outcome of this analysis is the
general view that Oenococcus is a genetically homogenous genus.
The strain sequenced
in this project, O. oeni PSU-1, was originally isolated at Penn
State University and is currently employed commercially to carry out
the malolactic fermentation wines.
References:
- Dellaglio, F.,
L. M. T. Dicks, and S. Torriani. 1995. The genus Leuconostoc, p.
235-278. In B. J. B. W. a. W. H. Holzapfel (ed.), The Genera of Lactic
Acid Bacteria, vol. 2. Blackie Academic & Professional, London.
- Dicks, L. M.,
F. Dellaglio, and M. D. Collins. 1995. Proposal to reclassify Leuconostoc
oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov.
International Journal of Systematic Bacteriology 45:395-7.
- Labarre, C.,
C. Diviès, and J. Guzzo. 1996. Genetic organization of the mle locus
and identification of a mleR-like gene from Leuconostoc
oenos. Applied and Environmental Microbiology 62:4493-8.
- Labarre, C.,
J. Guzzo, J. F. Cavin, and C. Diviès. 1996. Cloning and characterization
of the genes encoding the malolactic enzyme and the malate permease
of Leuconostoc oenos. Applied and Environmental Microbiology 62:1274-82.
- Morse, R., M.
D. Collins, K. Ohanlon, S. Wallbanks, and P. T. Richardson. 1996.
Analysis of the beta' subunit of DNA-dependent RNA polymerase does
not support the hypothesis inferred from 16S rRNA analysis that Oenococcus
oeni (formerly Leuconostoc oenos) is a tachytelic (fast-evolving)
bacterium. International Journal of Systematic Bacteriology 46:1004-1009.
- Versari, A.,
G. P. Parpinello, and M. Cattaneo. 1999. Leuconostoc oenos and
malolactic fermentation in wine: A review. Journal of Industrial
Microbiology & Biotechnology 23:447-455.
- Yang, D., and
C. R. Woese. 1989. Phylogenetic structure of the Leuconostocs: An
interesting case of a rapidly evolving organism. Systematic and Applied
Microbiology 12:145-149.
- Zuniga, M.,
I. Pardo, and S. Ferrer. 1996. Transposons Tn916 and Tn925 can transfer
from Enterococcus faecalis to Leuconostoc oenos. FEMS
Microbiology Letters 135:179-85.
|

