| |
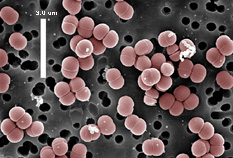
Photo: Jeff Broadbent, Utah State University |
Pediococcus pentosaceus are
Gram-positive, facultatively anaerobic, non-motile and non-spore-forming,
members of the industrially important lactic acid bacteria. Like other
lactic acid bacteria, P. pentosaceus are acid tolerant, cannot synthesize
porphyrins, and possess a strictly fermentative metabolism with lactic
acid as the major metabolic end product (Axelsson, 1998; Garvie, 1986).
Phylogenetically Pediococcus and Lactobacillus form a super-cluster
that can be divided in to two sub-clusters, all species of Pediococcus fall
within the Lactobacillus casei – Pediococcus sub-cluster. Morphologically,
pediococci (cocci; 0.6-1.0 mm in diameter) and lactobacilli (rods) are
distinct. The formation of tetrads via cell division in two perpendicular
directions in a single plane is a distinctive characteristic of pediococci. Pediococcus can
be described as “the only acidophilic, homofermentative, lactic acid
bacteria that divide alternatively in two perpendicular directions to form
tetrads” (Simpson and Taguchi, 1995). Lactic acid is produced from
hexose sugars via the Embden-Meyerhof pathway and from pentoses by the
6-phosphogluconate/phosphoketolase pathway (Axelsson, 1998). P. pentosaceus grow
at 40 but not 50oC, between pH 4.5 an 8.0, in 9-10% NaCl, hydrolyzes arginine,
can utilize maltose and some strains produce a “pseudo-catalase” (Garvie,
1986, Simpson and Taguchi, 1995).
Strains of P.
pentosaceus have been reported to contain between three and five
resident plasmids (Graham and McKay, 1985). Plasmid-linked traits
include the ability to ferment raffinose, melibiose, and sucrose,
as well as, the production of bacteriocins (Daeschel and Klaenhammer,
1985;Gonzalez and Kunka, 1986). Plasmids can be conjugally transferred
between Pediococcus and Enterococcus, Streptococcus,
or Lactococcus (Gonzalez and Kunka, 1983). Electroporation
has been utilized to introduce plasmids into pediococci, including
P. pentosaceus (Kim et al, 1992; Caldwell, 1996).
P. pentosaceus can
be isolated from a variety of plant materials and bacterial ripened
cheeses. This organism is used as an acid producing starter culture
in sausage fermentations, cucumber and green bean fermentations, soya
milk fermentations, and silage (Simpson and Taguchi, 1995). P. pentosaceus are
also a typical component of the adventitious or non-starter microflora
of most cheese varieties during ripening (Beresford et al., 2001).
In addition, it has been suggested that this organism may have value
as an acid-producing starter culture in the dairy fermentations (Caldwell
et al, 1996 and 1998).
Genetic studies
of P. pentosaceus have generated a limited quantity of information
(1 plasmid sequenced and 8 unique chromosomal sequences) on plasmid
and chromosomal encoded genes; however, the vast majority of genes
encoding industrially important attributes have yet to be described.
Genomic sequence analysis of P. pentosaceus genome will help
fill key knowledge gaps by providing a comprehensive view of the enzymes
and metabolic pathways related to: 1) acid and flavor production in
fermented meat and vegetable foods; 2) mechanisms by which by P.
pentosaceus and other nonstarter LAB grow and direct flavor development
in ripening cheese; and 3) mechanisms by which P. pentosaceus and
related lactic acid bacteria spoil wine and other alcoholic beverages.
In addition, improved knowledge of global gene regulation and integrative
metabolism in P. pentosaceus will also help to identify rational
strategies for metabolic and genetic improvements to industrial strains
of lactic acid bacteria.
References:
- Axelsson, L.
1998. Lactic acid bacteria: classification and physiology, pp. 1-72.
In, S. Salminen and A. Von Wright (eds). Lactic Acid Bacteria: Microbiology
and Functional Aspects, 2nd ed. Marcel Dekker, Inc, New York.
- Beresford, T.P.,
N.A. Fitzsimons, N.L. Brennan, T.M. Cogan. 2001. Recent advances
in cheese microbiology. Int. Dairy J. 11:259-274.
- Caldwell, S.,
D.J. McMahon, C.J. Oberg, and J.R. Broadbent. 1996. Development and
characterization of lactose-positive Pediococcus species for
milk fermentation. Appl. Environ. Microbiol. 62:936-941.
- Caldwell, S.,
R.W. Hutkins, DJ McMahon, CJ Oberg, and J.R. Broadbent. 1998. Lactose
and galactose uptake by genetically engineered Pediococcus species.
Appl. Microbiol. Biotechnol. 49:315-320.
- Daeschel, M.A.
and T.R. Klaenhammer. 1985. Association of a 13.6-megadalton plasmid
in Pediococcus pentosaceus with bacteriocin activity. Appl.
Environ. Microbiol. 50:1528-1541.
- Garvie, E.I.
1986. Genus Pediococcus, pp. 1075-1079. In, P. H. A. Sneath,
N. S. Mair, M. E. Sharpe, and J. G. Holt (eds.), Bergey's Manual
of Systematic Bacteriology, vol 2, 9th ed. Williams and Wilkins,
Baltimore.
- Gonzalez, C.F.
and B.S. Kunka. 1983. Plasmid transfer in Pediococcus spp.:
Intergeneric and intrageneric transfer of pIP501. Appl. Environ.
Microbiol. 46:81-89.
- Gonzalez, C.F.
and BS Kunka. 1986. Evidence for plasmid linkage of raffinose utilization
and associated a-galactosidase and sucrose hydrolase activity in Pediococcus
pentosaceus. Appl. Environ. Microbiol. 51:105-109.
- Graham, D.C.
and L.L. McKay. 1985. Plasmid DNA in strains of Pediococcus cerevisiae and Pediococcus
pentosaceus. Appl. Environ. Microbiol. 50:532-534.
- Kim, W.J., B.
Ray, and M.C. Johnson. 1992. Plasmid transfers by conjugation and
electroporation in Pediococcus acidilactici. J. Appl. Bacteriol.
72:201-207.
- Simpson W.J.
and H. Taguchi. 1995. The genus Pediococcus, with notes on
the genera Tetratogenococcus and Aerococcus, pp. 125-172.
In, B.J.B. Wood and W.H. Holzapfel (eds). The Genera of Lactic Acid
Bacteria. Chapman & Hall, London.
|

