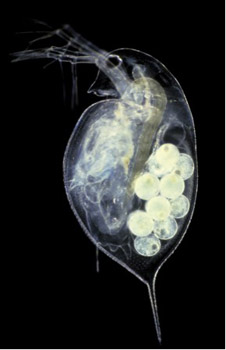
The globally distributed zooplankton Daphnia (commonly called the waterflea) is the first crustacean to have its genome sequenced. Daphnia has fascinated biologists for centuries because of its importance in aquatic ecosystems, its amenability to both field and laboratory study, and because of its remarkable ability and flexibility to cope with environmental challenges. These new genomic data will enhance studies in the wide variety of disciplines that make use of Daphnia for research - including crustacean biology, ecology, physiology, toxicology, population genetics, and evolution - and will promote greater understanding of the complex interplay between genome structure, gene expression, individual fitness, and population-level responses to environmental change. The availability of a Daphnia genome sequence will help create a new model system for ecological and evolutionary genomics.
There are numerous reasons why Daphnia is chosen for genomic study.
- Daphnia is a keystone species in both ponds and lakes. They are typically the principal grazers of algae, bacteria, protozoa, and the primary forage of fish. Because of their pivotal position in food webs, they are widely utilized as an indicator species to assess the response of ecosystems to environmental change.
- Daphnia has been used for decades as a standard organism for toxicity testing, and its toxicological reactions to environmental pollutants are well characterized. Of the nearly 500,000 records in the ECOTOX database, waterfleas represent 8% of all experimental data for aquatic organisms - second only to the rainbow trout.
- Daphnia is already a widely used model system for quantitative and ecological genetics.
- The reproductive cycle of Daphnia is ideal for experimental genetics. Generation time in the lab ranges from 5 - 10 days, making it possible to examine genome regulatory changes throughout its ontogeny.
- Distinct lineages of Daphnia have independently colonized radically different environments (deep permanent lakes vs. shallow temporary ponds) on multiple occasions. These evolutionarily independent radiations provide an opportunity to evaluate whether similar environmental challenges are countered in genetically consistent ways.
- Daphnia is a natural outgroup for comparative genomic studies among the arthropods, by appropriately rooting the phylogenetic tree of the model insect species. Crustaceans and insects have diverged from a common ancestor; therefore genomic characteristics are either derived or preserved, in reference to the Daphnia genome.
The genome sequence is supported by an international research community that is building additional resources for its annotation and functional characterization, including genomic and cDNA libraries, an EST database of over 200,000 sequences, genetic linkage maps for its 12 chromosomes, microarrays for gene expression studies, a protein sequence database, and a web-based bioinformatics portal for public access to the data. The Daphnia Genomics Consortium and the JGI are coordinating a community-wide analysis of the data. Version 1.0 of the D. pulex genome sequence assembly and annotation is a work in progress. Investigators who are interested in participating can visit the consortium webpage for information. The clone that was sequenced, "The Chosen One", is available from the CGB.
Genome Reference(s)
Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Cáceres CE, Carmel L, Casola C, Choi JH, Detter JC, Dong Q, Dusheyko S, Eads BD, Fröhlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kültz D, Laforsch C, Lindquist E, Lopez J, Manak JR, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Koonin EV, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL
The ecoresponsive genome of Daphnia pulex.
Science. 2011 Feb 4;331(6017):555-61. doi: 10.1126/science.1197761
